Project Details
Project Description
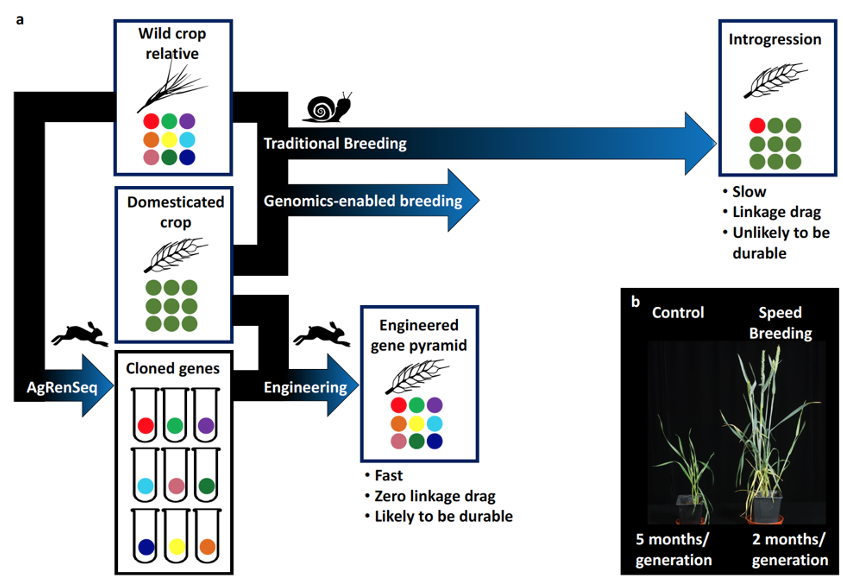
Wheat rusts are destructive diseases of wheat, which throughout recorded history have caused devastating epidemics almost wherever wheat is grown. The wild ancestors of domesticated wheat represent a rich source of genetic variation with huge potential for improving disease resistance. Deploying this genetic diversity into elite, cultivated wheat by traditional breeding takes many years for just a single resistance gene. However, the molecular identification (cloning) of resistance genes opens up new possibilities for accelerated breeding by marker-assisted selection and genetic engineering [Refs. 1,2]. The Wulff lab has established a suite of molecular plant breeding technologies that significantly reduce costs and accelerate plant growth [Ref. 3], gene discovery, and gene cloning [Refs. 4-9]. You will use our tools, structured germplasm, and sequence resources to characterize novel candidate rust resistance genes. Alternatively, you may chose to study the mechanism of non-cannonical resistance genes [ref. 10], or support our efforts to engineer wheat plants with improved diseae resistance [11]. You will be supervised by Brande Wulff and receive training and co-supervision from a team of Postdocs and PhD students with expertise in bioinformatics, mathematics, scripting, genetics, plant pathogen interactions, wheat husbandry and crossing. KAUST is a vibrant place to discuss and plan science. You will become part of the larger KAUST community and alumni, which we hope will have lasting positive impacts on your future career.
References [1] Dhugga & Wulff (2018). Science 361:451-452. [2] Luo et al (2021) Nature Biotechnology 39:561-566. [3] Watson et al (2017) Nature Plants 4:23-29. [4] Sánchez-Martín et al (2016). Genome Biology 17:221. [5] Steuernagel et al (2016). Nature Biotechnology 34:652-5. [6] Arora et al (2018) Nature Biotechnology 2:139-143. [7] Steuernagel et al (2020) Plant Physiology 183:468-482. [8] Gaurav et al. (2022) Nature Biotechnology 40:422–431. [9] Cavalet-Giorsa et al (2024) Nature 633:848–855. [10] Chen et al (2025) Science 387:1402-1408 [11] Ayala et al (2024) Annual Review of Phytopathology 62:193-215.
About the Researcher
Affiliations
Education Profile
- Postdoc, IBMP-CNRS, Strasbourg, France, 2008
- Human Frontiers Postdoc Fellow, IBMCP-CSIC, Spain, 2005
- EMBO Postdoc Fellow, IBMCP-CSIC, Spain, 2004
- PhD, The Sainsbury Laboratory, UK, 2001
- BSc, University of East Anglia, UK, 1997
Research Interests
a€‹Brande's research programme explores the genetics of disease resistance in wheat. This has led to developing fast, new and efficient methods for gene discovery and cloning which use mutant and natural populations followed by sequence alignment to locate genes. Brande has also co-developed a method for halving the generation time of wheat and other crops, in a controlled environment, dramatically speeding up capabilities for research and breeding purposes. His long-term aim is to use cloned genes from wild ancestors of wheat to engineer durable resistance to these diseases in cultivated wheat.Selected Publications
- Reference genome-assisted identification of stem rust resistance gene Sr62 encoding a tandem kinase, Yu G, Matny O, Champouret N, Steuernagel B, Moscou MJ, HernA¡ndez-PinzA³n I, Green P, Hayta S, Smedley M, Harwood W, Kangara N, Yue Y, Gardener C, Manfield MJ, Olivera PD, Welchin C, Simmons J, Millet E, Minz-Dub A, Ronen M, Avni R, Sharon A, Patpour M, Justesen Af, Jayakodi M, Himmelbach A, Stein N, Wu S, Poland J, Ens J, Pozniak C, KarafiA¡tovA¡ M, MolnA¡r I, DoleA¾el J, Ward ER, Reuber TL, Jones JDG, Mascher M, Steffenson BJ, Wulff BBH, Nature Communications, 2022, 13:1607.
- Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement, Gaurav K, Arora S, Silva P, SA¡nchez-MartAn J, Horsnell R, Gao L, Brar GS, Widrig V, Raupp J, Singh N, Wu S, Kale Sm, Chinoy C, Nicholson P, Quiroz-ChA¡vez J, Simmonds J, Hayta S, Smedley MA, Harwood W, Pearce S, Gilbert D, Kangara N, Gardener C, Forner-MartAnez M, Liu J, Yu G, Boden S, Pascucci A, Ghosh S, Hafeez An, O'hara T, Waites J, Cheema J, Steuernagel B, Patpour M, Justesen AF, Liu S, Rudd JC, Avni R, Sharon A, Steiner B, Pkirana RP, Buerstmayr H, Mehrabi AA, Nasyrova FY, Chayut N, Matny O, Steffenson BJ, Sandhu N, Chhuneja P, Lagudah E, Elkot AF, Tyrrell S, Bian X, Davey RP, Simonsen M, Schauser L, Tiwari VK, Kutcher HR, Hucl P, Li A, Liu D-C, Mao L, Xu S, Brown-Guedira G, Faris J, Dvorak J, Luo M-C, Krasileva K, Lux T, Artmeier S, Mayer KFX, Uauy C, Mascher M, Bentley AR, Keller B, Poland J, Wulff BBH, Nature Biotechnology, 2022, 40:422a-431.
- Creation and judicious application of a wheat resistance gene atlas, Hafeez AN, Arora S, Ghosh S, Gilbert D, Bowden RL, Wulff BBH, Molecular Plant, 2021, 14:1053a-1070.
- Breeding a fungal gene into wheat, WULFF BBH, JONES JDG, Science, 2020, 368:822-823.
- Resistance gene cloning from a wild crop relative by sequence capture and association genetics, Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O, Johnson R, Enk J, Periyannan S, Narinder S, Hatta MAMD, Athiyannan N, Cheema J, Yu G, Kangara N, Ghosh S, Szabo LJ, Poland J, Bariana H, Jones JDG, Bentley AR, Ayliffe M, Olson E, Xu SS, Steffenson BJ, Lagudah E, Wulff BBH, Nature Biotechnology, 2019, 37:139-143.
Desired Project Deliverables
Recommended Student Background
We are shaping the
World of Research
Be part of the journey with VSRP